MedML: Fusing Medical Knowledge and Machine Learning Models for Early Pediatric COVID-19 Hospitalization and Severity Prediction
Abstract
Background: The COVID-19 pandemic has caused devastating economic and social disruption, straining the resources of healthcare institutions worldwide. The virus continues to evolve rapidly, and the arrival of each new strain brings with it a real threat of resurgence. This has led to a nationwide call for models to predict hospitalization and severe illness in patients with COVID-19 to inform distribution of limited healthcare resources. We respond to one of these calls specific to the pediatric population.
Tasks: To address this challenge, we study two prediction tasks for the pediatric population using electronic health records: 1) predicting which children are more likely to be hospitalized, and 2) among hospitalized children, which individuals are more likely to develop severe symptoms.
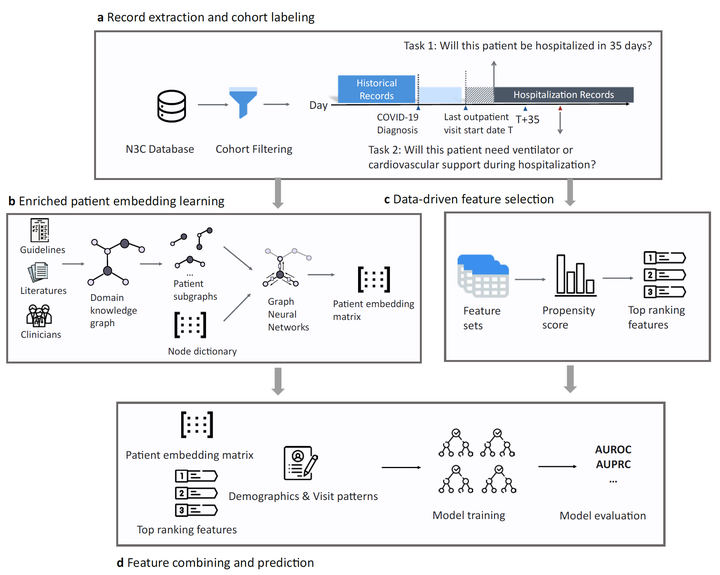
Methodology: We respond to the national Pediatric COVID-19 data challenge with a novel machine learning model, MedML. MedML extracts the most predictive features based on medical knowledge and propensity scores from over 6 million medical concepts and incorporates the inter-feature relationships between heterogeneous medical features, including measurements, medications, conditions, and procedures in the form of medical knowledge graphs via graph neural networks (GNN). The final predictions are computed using ensemble models with learned patient graph embeddings and feature vectors. We evaluate MedML across 143,605 patients for the hospitalization prediction task and 11,465 patients for the severity prediction task using data from the National Cohort Collaborative (N3C) dataset, a multi-site large-scale heterogeneous collection of Electronic Health Record (EHR) data available to the public. We also report detailed group-level and individual-level feature importance analyses to evaluate the model interpretability.
Result: MedML achieves 0.75 area under receiver operating characteristic curve (AUROC) and 0.15 area under the precision-recall curve (AUPRC) score for the hopitalization prediction; 0.69 AUROC and 0.22 AUPRC for the severity prediction. MedML achieves up to a 7% higher AUROC score and up to a 14% higher AUPRC score compared to the best baseline machine learning models and performs well across all nine national geographic regionsand over all three-month spans since the start of the pandemic.
Conclusion: Our cross-disciplinary research team has developed a method of incorporating clinical domain knowledge as the framework for a new type of machine learning model that is more predictive and explainable than current state-of-the-art data-driven feature selection methods.